About pirScan
Motivation of pirScan
C.elegans is one of the most wildly used model organisms. However, the expression of transgenes in the germline of C.elegans is notoriously difficult due to PIWI interacting RNA (piRNA)-mediated gene silencing. While several web tools are available to predict miRNA targets, a similar tool is currently unavailable for piRNA. Using systematic piRNA reporter assays, we and others have demonstrated that piRNAs prefer perfect base pairing at a seed region but otherwise can tolerate a few mismatches. Here we develop the pirScan webtool to predict the targeting sites of 15,000 piRNAs produced in C.elegans and thereby avoid transgene silencing. To validate that pirScan can predict piRNA sites successfully, we predicted piRNA sites on three silencing-prone transgenes, which carry GFP, mCherry and Cas9, respectively. Remarkably, by introducing silent mutations to remove piRNA targeting sites, we observed expression of all three transgenes in the germline of C.elegans.
Impact of pirScan
pirScan provides the first webserver that predicts piRNA targeting sites. We expect pirScan to hugely impact the C.elegans research community: not only by providing researchers with a platform to examine piRNA targeting sites in a given sequence, but also by allowing researchers to circumvent the transgene silencing that has confounded the field for two decades.
General Framework
pirScan is a tool that predicts the presence of piRNA targeting sites within a given sequence and suggests silent mutations that can be introduced to avoid silencing of transgenes by piRNAs. The user can input a mature mRNA or spliced DNA sequence in plain text format and be provided with a concise graphical representation of which sites within the sequence display predicted piRNA targeting sites. The user can then choose between suggested silent mutations that will allow the input sequence to escape predicted piRNA targeting, but still retain the same amino acid sequence.
For each piRNA targeting site, pirScan lists the types and positions of mismatches between a predicted endogenous piRNA and the user’s input sequence. The pairing between the 21 nt long piRNA and the corresponding position in the input sequence is displayed for clarity, with the input sequence listed 5’ to 3’. The target sites within the input sequence can be downloaded as a .csv table or as a graphic to be displayed or manipulated by the user outside of our provided framework.
A user can subsequently change his/her input sequence by following our intuitive graphical interface. Proposed silent mutations will be displayed which can be chosen by the user. Each proposed mutation also indicates which targeting rule(s) will be broken if the nucleotide is changed. The proposed changes are ranked so that mutations which break the most important rules (i.e. mismatches within the seed region) will be listed first and selected by default. If more than one piRNA targeting site were present in the input sequence, then each piRNA will have its own separate table complete with the possible silent mutations to introduce. Each table is displayed according to the 5’ to 3’ order which each piRNA is predicted to interact with the input sequence. Upon the user’s completion of the proposed mutagenesis, the modified sequence can be immediately re-submitted for scanning to verify that the sequence will successfully escape piRNA targeting and ensure that new targeting sites were not inadvertently introduced. If pirScan still predicts piRNA targeting sites in the modified sequence, the suggested changes for these sites are presented for further modification. Once the user is satisfied with the adjusted input sequence, the modified version can be downloaded as a text file. Additionally, a table and graphic which highlights the user-selected mutations are available for export.
Input
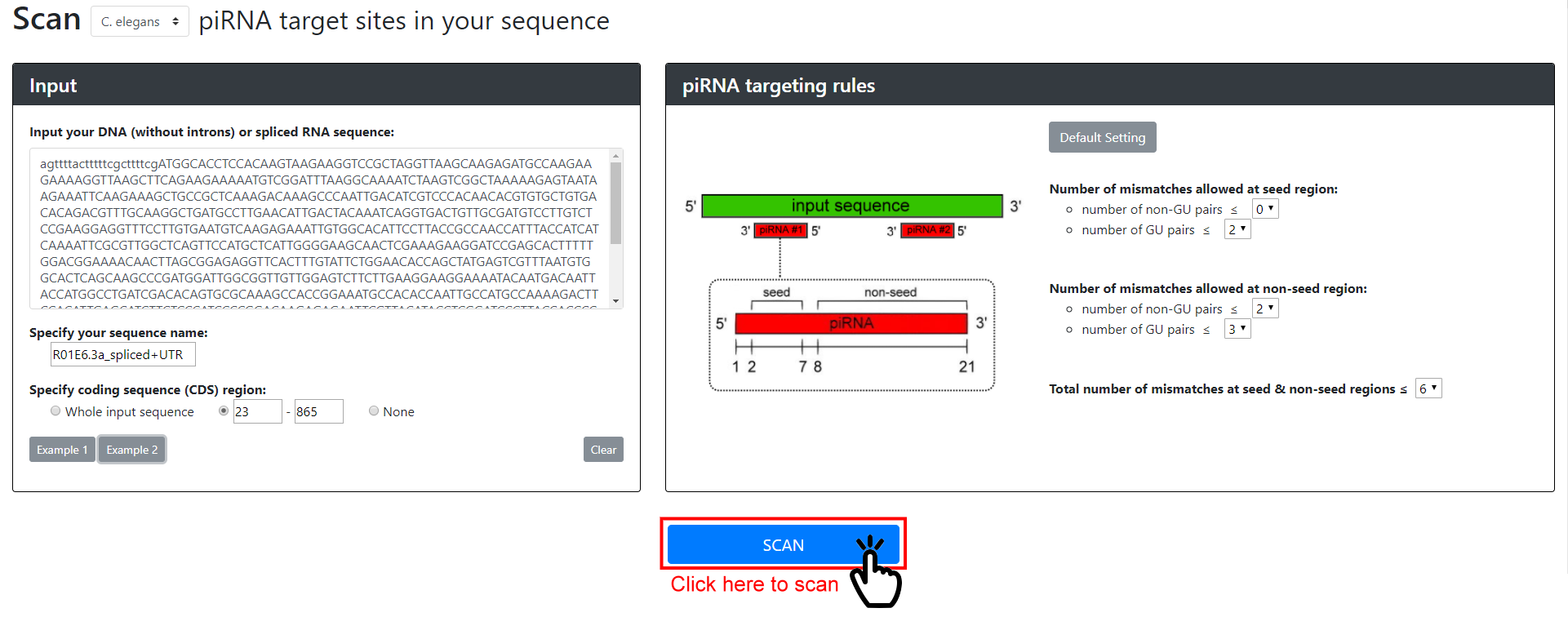
pirScan will accept an input sequence in plain text format. The user may also give the input a name, which will be included in the output following modifications. The input can be a mature mRNA sequence or spliced DNA sequence. By default, pirScan will assume that the first nucleotide present is the start of the open reading frame. If the first nucleotide is not the start of the open reading users can also specify the numerical positions of the open reading frame to provide coding information of the input sequence by changing the selection in the input window . As piRNAs can also target 5’ and 3’ UTR, we suggest the user to include the sequence of 5’ and/or 3’ UTRs and simply specify the coding region. The input sequence should not contain introns. ‘None’ can be selected as the coding sequence region.
piRNA Targeting Rules
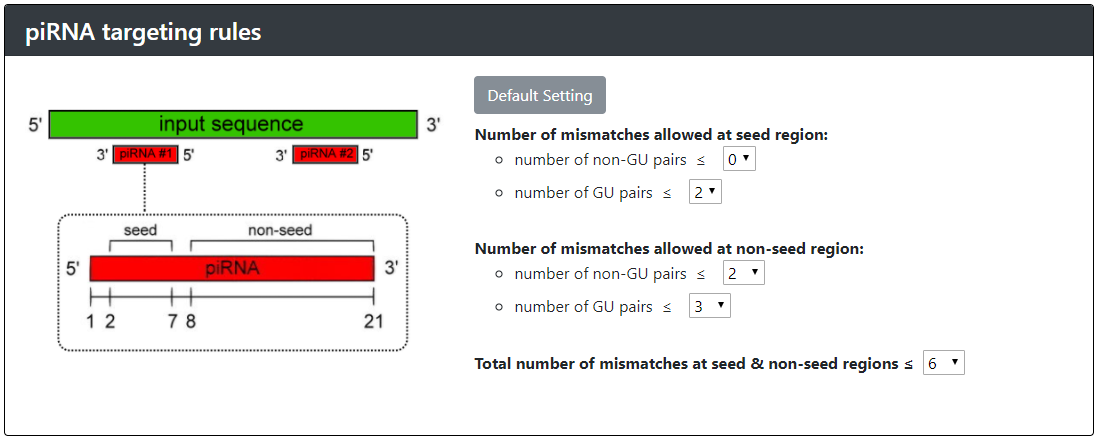
The rules governing piRNA targeting have proven enigmatic. It has been shown, however, that piRNAs can tolerate at least three mismatches between their sequence and a target. Our group has resolved the mystery by examining how a single piRNA recognizes its target, and confirming our observations by engineering synthetic piRNAs in a reporter-based assay. These analyses have revealed that piRNAs require perfect matching within the 2nd to 7th nucleotide seed-region and can only tolerate a few mismatches outside of the seed region but the first nucleotide does not contribute to the piRNA targeting. pirScan will identify piRNA sites using the default setting if not specified. The default setting reflects the confident piRNA targeting sites according to our reporter assay.
piRNA Target Site Identification Output
Upon submission of an input sequence, the user can download a table in .csv format that contains the list of piRNAs predicted to target the input sequence, the targeting score, the location of targeting sites, how many mismatches are present, and some aspects about those mismatches. The targeting score has a maximum of 10, and any targets with a score of 0 or above should be considered as possibly sufficient for silencing. Additionally, a visual representation of the predicted piRNA targeting to the input sequence is available for downloading in .png format. Either of these outputs can allow a user to customize his/her input sequence outside of our pirScan framework is he/she chooses.

Modification of the Input Sequence to Avoid piRNA Targeting
At the end of the output page of piRNA targets sites, a link is offered for users to introduce silent mutations in their input sequences to avoid piRNA recognition. The system lists up to 6 suggested changes, ranked by the targeting score from highest to lowest. The system will by default choose the top suggested change for the user but the user can select/deselect any modifications or select multiple modifications at each piRNA sites. The system also displays which rule(s) each mutation will break. Note that sometimes multiple mutations are needed for avoiding piRNA recognition. Importantly, pirScan will not suggest mutations that will result in the incorporation of a rarely used codon.
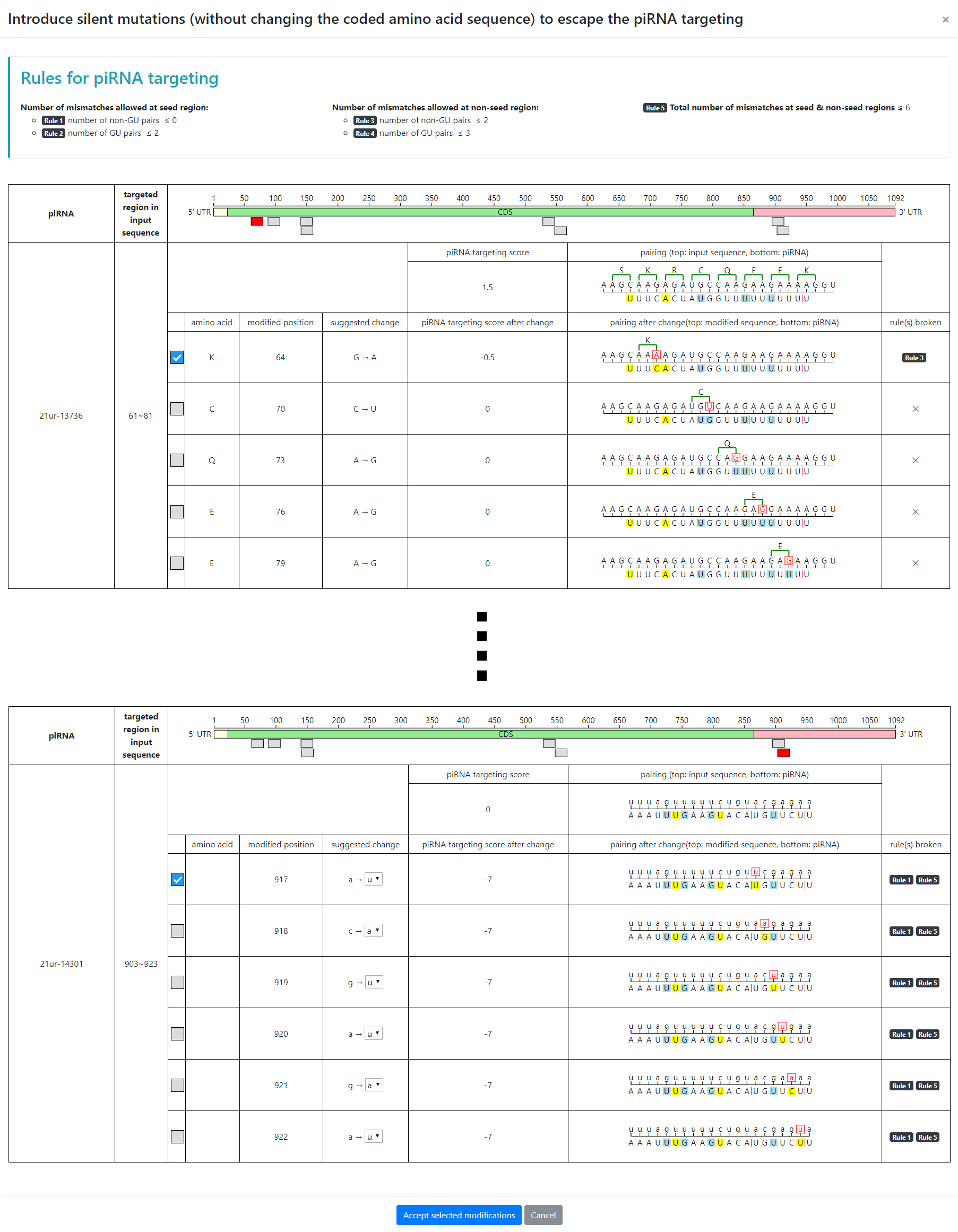
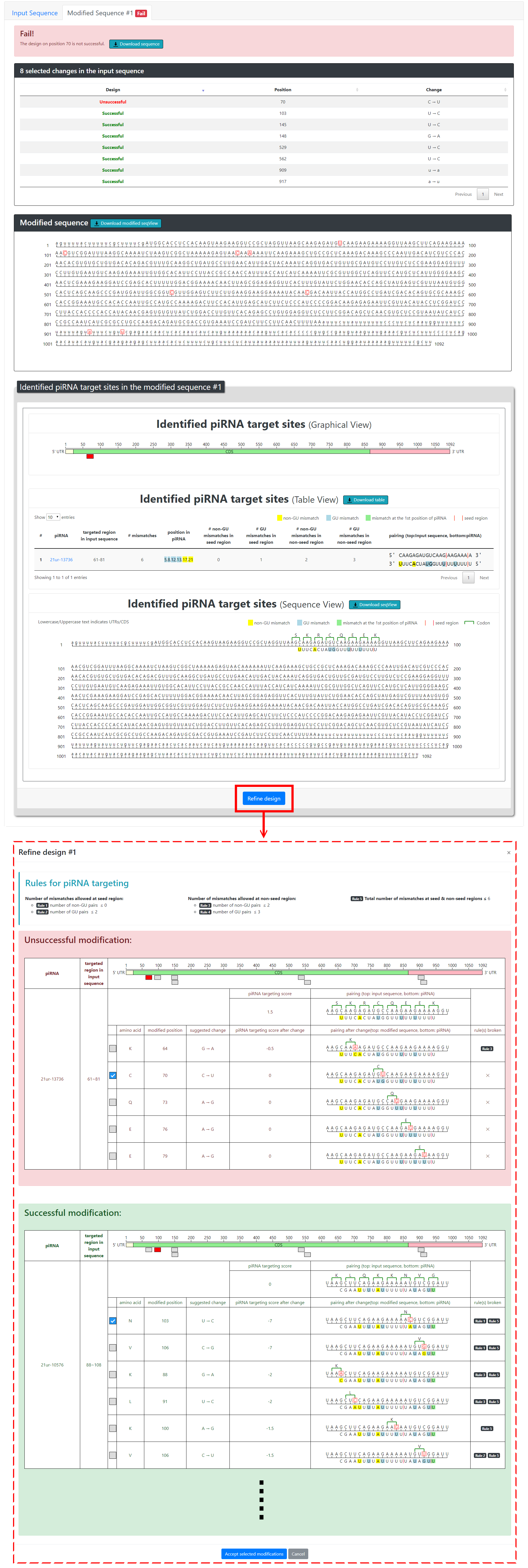
Once the user chooses silent mutations to introduce into the input sequence, the program will display a table summarizing the changes and also automatically submit the modified sequence for re-scanning. pirScan will then display “Success” if no piRNA targeting sites are found in the modified sequence, or “Fail” if additional piRNA targeting sites are found. The modified sequences (either success or failure) can be downloaded in plain text format.
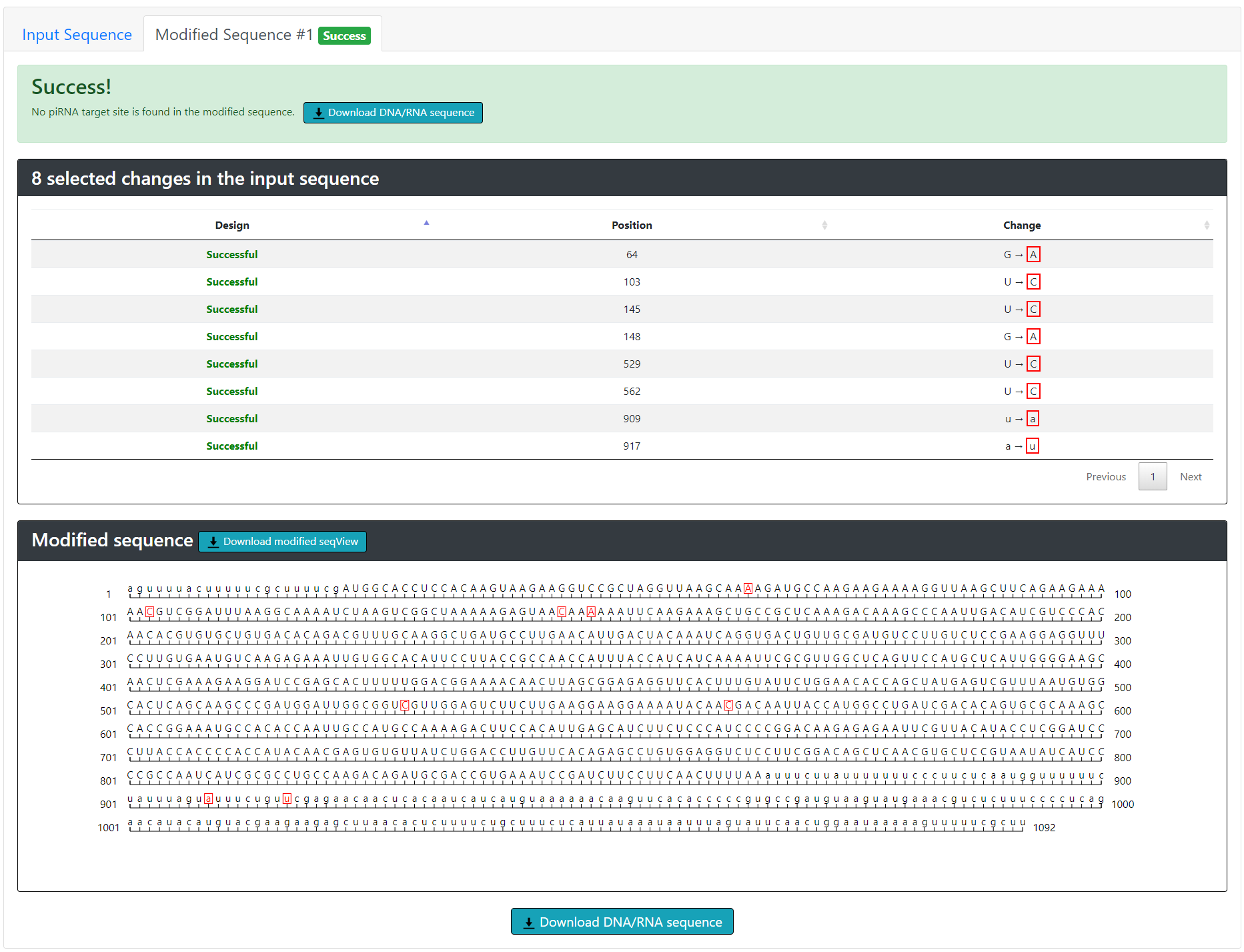
In the case of failure, the program first displays the piRNA sites that remain to allow further modification. We suggest to the user to choose additional sites if a single mutation is not sufficient to avoid piRNA recognition, or to choose different sites if a new piRNA targeting site is found.