Scan piRNA target sites in your sequence
Input
Specify your sequence name:
Specify coding sequence (CDS) region:
piRNA targeting rules
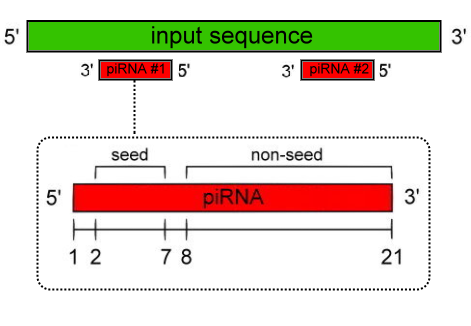
- Number of mismatches allowed at seed region:
-
- Number of mismatches allowed at non-seed region:
-
To examine piRNA sites in endogenous genes and to evaluate their effects in gene silencing, please visit piRTarBase.
Reference
If you make use of the pirScan webserver presented here, please cite the following articles:
- Wu WS; Huang WC; Brown J; Zhang D; Song X; Chen H; Tu S; Weng Z; Lee HC, 2018 “pirScan, a webserver to predict piRNA targeting sites and to avoid transgene silencing in C. elegans” Nucleic Acids Res. doi: 10.1093/nar/gky277
- Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, Lee HC. 2018 “The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes”, Science. 359, 587-592