What is YHMI?
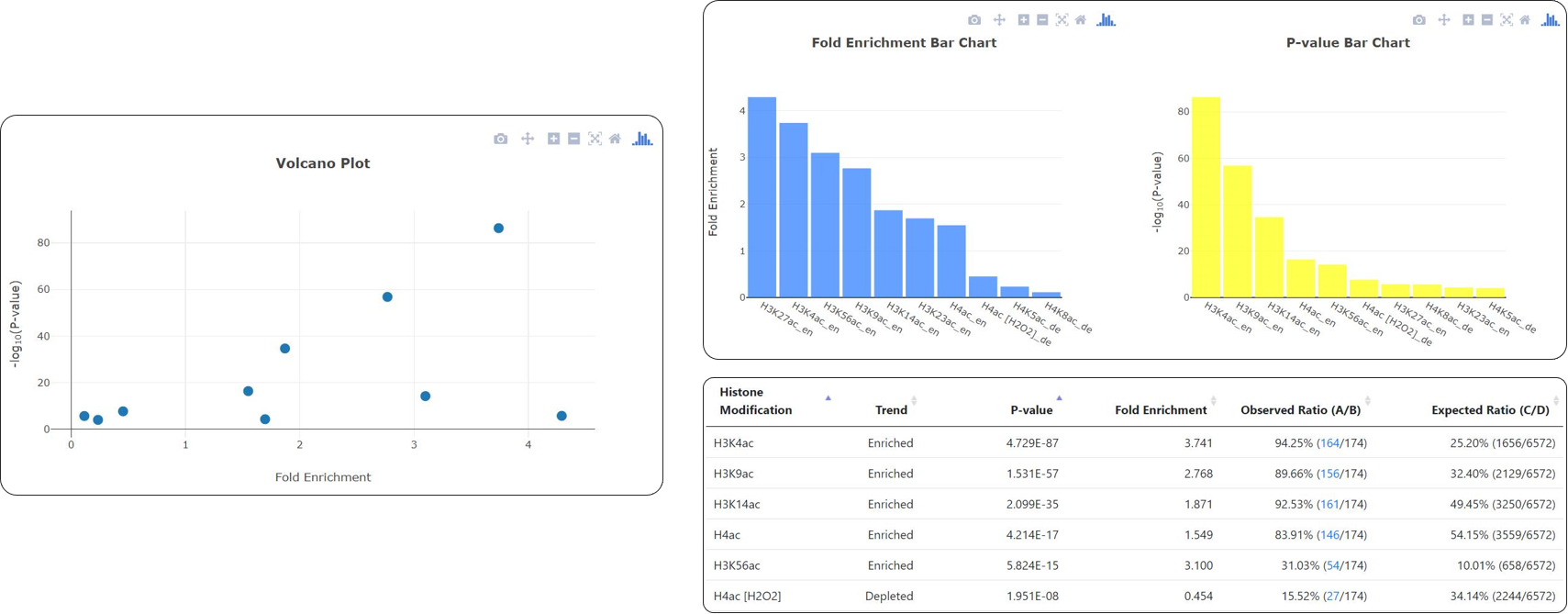
YHMI is a web tool to identify the enriched/depleted histone modifications and the enriched histone/chromatin regulators for a gene list.
YHMI uses ChIP-chip/ChIP-seq datasets of 32 histone modifications and 83 histone/chromatin regulators.
15 histone acetylation H2AK5ac, H3K4ac, H3K9ac, H3K14ac, H3K14ac [H2O2], H3K18ac, H3K23ac, H3K27ac, H3K56ac, H4ac, H4ac [H2O2], H4K5ac, H4K8ac, H4K12ac, H4K16ac 13 histone methylation H3R2me2a, H3K4me, H3K4me2, H3K4me3, H3K36me, H3K36me2, H3K36me3, H3K79me, H3K79me2, H3K79me3, H4R3me, H4R3me2s, H4K20me 2 histone phosphorylation H2AS129ph, H3S10ph 1 histone ubiquitination H2BK123ub 1 histone variant H2AZ 83 histone/chromatin regulators (in 25℃ and 37℃) Ada2, Brn1, Esa1, Gcn5, Rsc4, Rxt2, Snf1, Swc1, Yaf9, ... See more. -
The identification results are shown both in figures and tables.
Two Related Websites
Saccharomyces Genome Database (SGD) comprehensively collects the yeast histone modification datasets from the literature and allows users to visualize various histone modifications using JBrowse (a genome browser). [Example]
The Yeast Nucleosome Atlas (YNA) implements a tool for users to retrieve a list of yeast genes whose promoters and/or coding regions contain a specific combination of histone modifications (e.g. H3K4ac & H3K4me3 in promoters).
Update History
| 2018 Aug. | Provide volcano plots to visualize the identification results. |
| 2018 July | Include ChIP-seq datasets of 17 histone modifications (9 histone acetylation, 6 hsitone methylation and 2 histone phosphorylation). |
| 2018 June | Release YHMI. |
| 2018 Jan. | Construct the web interface of YHMI. |
| 2017 Nov. | Include ChIP-chip datasets of 15 histone modifications (6 histone acetylation, 7 histone methylation, 1 histone ubiquitination and 1 histone variant). |
| 2017 Sept. | Start YHMI project. |