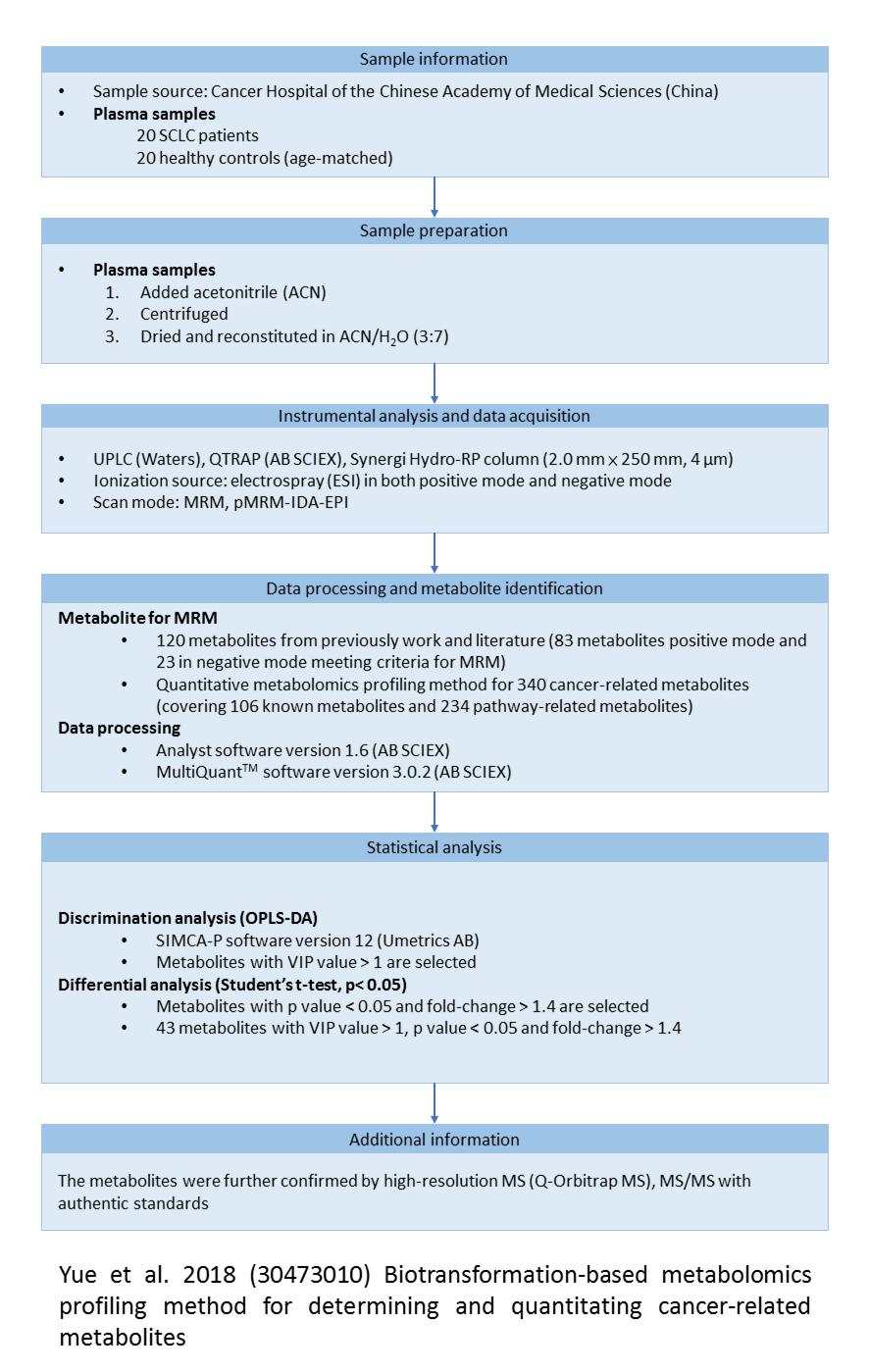
Showing paper detailed information
Citation Information
J Pharm Biomed Anal. 2019 Dec 23;180:113069. doi: 10.1016/j.jpba.2019.113069. [Epub ahead of print]
Metabolic and lipidomic characterization of malignant pleural effusion in human lung cancer.
Yang Z, Song Z, Chen Z, Guo Z, Jin H, Ding C, Hong Y, Cai Z.
Analytical methods | ||
|---|---|---|
| Comparative study | #1 | |
| Chromatography | LC | |
| Ion source | ESI | |
| Positive/Negative mode | positive, negative | |
| Mass analyzer | Q-Orbitrap | |
| Identification level | MS/MS | |
Sample information | ||
|---|---|---|
| Comparative study | #1 | |
| Country | China | |
| Specimen | pleural effusion | |
| Marker function | diagnosis | |
| Participants(Case) | Cancer type | adenocarcinoma |
| Stage | – | |
| Number | 46 | |
| Gender (M,F) | 15, 31 | |
| Mean age (range) (M,F) | 63 ± 12 | |
| Smoking status | – | |
| Participants(Control) | Type | pulmonary tuberculosis, other pulmonary diseases |
| Number | 32.0 | |
| Gender (M,F) | 26, 6 | |
| Mean age (range) (M,F) | 49 ± 19 | |
| Smoking status | – | |
Data processing and metabolite identification | |
|---|---|
| Data processing software | XCMS |
| Database search | HMDB, METLIN, LipidSearch |
Statistics and concentration information | |
|---|---|
| Differential analysis method | PLS-DA |
| Classification method | ROC analysis |
| Survival analysis method | – |
Metabolites identified in the paper
| Metabolite | Comparative study | Author-emphasized biomarkers | Mean concentration (case) | Mean concentration (control) | Fold change (case/control) | P-value | FDR | VIP |
|---|---|---|---|---|---|---|---|---|
| 2-methylbutyroyl-carnitine | – | – | – | – | 0.54 | – | 0.00461 | 1.32 |
| arginine | – | – | – | – | 1.38 | – | 0.00009 | 1.04 |
| Cer(d18:1/16:0) | – | – | – | – | 0.65 | – | 0.00935 | 1.17 |
| Cer(d18:2/24:1) | – | – | – | – | 0.61 | – | 0.00000552 | 1.6 |
| ChE(16:1) | – | – | – | – | 1.59 | – | 0.00369 | 1.18 |
| ChE(18:3) | – | – | – | – | 1.49 | – | 0.00197 | 1.1 |
| citric acid | – | – | – | – | 1.53 | – | 0.00882 | 1.24 |
| cortisol | – | – | – | – | 0.43 | – | 0.0372 | 1.42 |
| DiHOME | – | – | – | – | 3.24 | – | 0.0000805 | 2.35 |
| dodecanoyl-carnitine | – | – | – | – | 0.63 | – | 0.00776 | 1.19 |
| glutamate | – | – | – | – | 1.84 | – | 0.00012 | 1.69 |
| glycocholic acid | – | – | – | – | 0.3 | – | 0.047 | 1.52 |
| HODE | – | – | – | – | 3.08 | – | 0.00449 | 1.94 |
| HOTrE | – | – | – | – | 3.5 | – | 0.0000161 | 3.21 |
| HpODE | – | – | – | – | 2.83 | – | 0.00127 | 2.81 |
| HpOTrE | – | – | – | – | 3.51 | – | 0.000526 | 3.02 |
| LPC(16:0e) | – | – | – | – | 0.38 | – | 0.000000284 | 2.09 |
| LPC(18:0e) | – | – | – | – | 0.41 | – | 0.000000000351 | 2.19 |
| LPC(18:0p) | – | – | – | – | 0.66 | – | 0.00352 | 1.2 |
| LPC(20:0e) | – | – | – | – | 0.5 | – | 0.000000533 | 1.95 |
| LPC(20:2) | – | – | – | – | 0.52 | – | 0.0000319 | 1.58 |
| LPC(20:4) | – | – | – | – | 0.56 | – | 0.00155 | 1.28 |
| LPC(26:0) | – | – | – | – | 0.6 | – | 0.0000496 | 1.52 |
| LPC(26:1) | – | – | – | – | 0.57 | – | 0.0000334 | 1.63 |
| myristoylcarnitine | – | – | – | – | 0.4 | – | 0.000388 | 1.74 |
| N1-Acetyl-spermidine | – | – | – | – | 0.7 | – | 0.0151 | 1.14 |
| octadecadienoic acid | – | – | – | – | 2.81 | – | 0.0000204 | 2.95 |
| OxoODE | – | – | – | – | 2.6 | – | 0.000302 | 2.93 |
| OxoOTrE | – | – | – | – | 2.63 | – | 0.000158 | 3.7 |
| PC(15:0/20:2) | – | – | – | – | 2.03 | – | 0.000238 | 1.73 |
| PC(16:0/22:4) | – | – | – | – | 1.47 | – | 0.00109 | 1.17 |
| PC(16:0e/20:4) | – | – | – | – | 0.67 | – | 0.0000000154 | 1.39 |
| PC(18:0e/22:4) | – | – | – | – | 0.51 | – | 0.00000000392 | 1.86 |
| PC(18:0p/22:5) | – | – | – | – | 0.67 | – | 0.0000548 | 1.17 |
| PC(18:1p/22:6) | – | – | – | – | 1.34 | – | 0.00935 | 1.1 |
| PC(19:0/18:2) | – | – | – | – | 1.23 | – | 0.00919 | 0.8 |
| PC(30:0e) | – | – | – | – | 0.59 | – | 0.0000227 | 1.56 |
| PC(33:0) | – | – | – | – | 0.68 | – | 0.00162 | 1.24 |
| PC(34:0e) | – | – | – | – | 0.59 | – | 0.00000166 | 1.68 |
| PC(34:2e) | – | – | – | – | 0.66 | – | 0.00000495 | 1.46 |
| PC(35:0) | – | – | – | – | 0.47 | – | 0.00162 | 1.63 |
| PC(36:0e) | – | – | – | – | 0.52 | – | 0.0000138 | 1.75 |
| PC(36:4e) | – | – | – | – | 0.67 | – | 0.0000000116 | 1.55 |
| PC(38:6e) | – | – | – | – | 0.56 | – | 0.00000821 | 1.7 |
| PC(40:3p) | – | – | – | – | 0.53 | – | 0.0000000000243 | 2.05 |
| PC(40:5e) | – | – | – | – | 0.48 | – | 0.000000917 | 1.97 |
| PC(40:6e) | – | – | – | – | 0.71 | – | 0.00026 | 1.2 |
| PC(40:7p) | – | – | – | – | 0.63 | – | 0.000343 | 1.38 |
| PC(42:1) | – | – | – | – | 0.69 | – | 0.000601 | 1.21 |
| PC(42:3p) | – | – | – | – | 0.52 | – | 0.0000000552 | 1.94 |
| PC(44:6e) | – | – | – | – | 0.54 | – | 0.000000412 | 1.85 |
| PC(44:7e) | – | – | – | – | 0.58 | – | 0.0000478 | 1.56 |
| PE(16:0p/20:4) | – | – | – | – | 0.58 | – | 0.000000953 | 1.74 |
| PE(16:0p/22:4) | – | – | – | – | 0.4 | – | 0.00000227 | 2.11 |
| PE(18:0p/18:1) | – | – | – | – | 0.53 | – | 0.005 | 1.37 |
| PE(18:1p/18:1) | – | – | – | – | 0.62 | – | 0.00312 | 1.27 |
| phSM(d34:2) | – | – | – | – | 0.66 | – | 0.00000238 | 1.37 |
| phSM(d42:1) | – | – | – | – | 1.23 | – | 0.00684 | 0.72 |
| PI(18:0/20:3) | – | – | – | – | 1.52 | – | 0.000112 | 1.19 |
| SM(d32:0) | – | – | – | – | 0.63 | – | 0.000163 | 1.39 |
| TG(18:2/18:2/22:6) | – | – | – | – | 0.6 | – | 0.00011 | 1.54 |
| TG(18:3/18:2/20:4) | – | – | – | – | 0.71 | – | 0.0106 | 1.04 |
| Metabolite | Comparative study | Author-emphasized biomarkers | Cutoff value | AUROC (95%CI) | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| 2-methylbutyroyl-carnitine | – | – | – | 0.733 | – | – | – |
| arginine | – | – | – | 0.81 | – | – | – |
| Cer(d18:1/16:0) | – | – | – | 0.696 | – | – | – |
| Cer(d18:2/24:1) | – | – | – | 0.861 | – | – | – |
| ChE(16:1) | – | – | – | 0.73 | – | – | – |
| ChE(18:3) | – | – | – | 0.731 | – | – | – |
| citric acid | – | – | – | 0.728 | – | – | – |
| cortisol | – | – | – | 0.859 | – | – | – |
| DiHOME | – | – | – | 0.793 | – | – | – |
| dodecanoyl-carnitine | – | – | – | 0.744 | – | – | – |
| glutamate | – | – | – | 0.839 | – | – | – |
| glycocholic acid | – | – | – | 0.744 | – | – | – |
| HODE | – | – | – | 0.759 | – | – | – |
| HOTrE | – | – | – | 0.845 | – | – | – |
| HpODE | – | – | – | 0.787 | – | – | – |
| HpOTrE | – | – | – | 0.779 | – | – | – |
| LPC(16:0e) | – | – | – | 0.889 | – | – | – |
| LPC(18:0e) | – | – | – | 0.935 | – | – | – |
| LPC(18:0p) | – | – | – | 0.733 | – | – | – |
| LPC(20:0e) | – | – | – | 0.858 | – | – | – |
| LPC(20:2) | – | – | – | 0.793 | – | – | – |
| LPC(20:4) | – | – | – | 0.733 | – | – | – |
| LPC(26:0) | – | – | – | 0.813 | – | – | – |
| LPC(26:1) | – | – | – | 0.833 | – | – | – |
| myristoylcarnitine | – | – | – | 0.79 | – | – | – |
| N1-Acetyl-spermidine | – | – | – | 0.734 | – | – | – |
| octadecadienoic acid | – | – | – | 0.837 | – | – | – |
| OxoODE | – | – | – | 0.801 | – | – | – |
| OxoOTrE | – | – | – | 0.805 | – | – | – |
| PC(15:0/20:2) | – | – | – | 0.782 | – | – | – |
| PC(16:0/22:4) | – | – | – | 0.738 | – | – | – |
| PC(16:0e/20:4) | – | – | – | 0.892 | – | – | – |
| PC(18:0e/22:4) | – | – | – | 0.892 | – | – | – |
| PC(18:0p/22:5) | – | – | – | 0.782 | – | – | – |
| PC(18:1p/22:6) | – | – | – | 0.715 | – | – | – |
| PC(19:0/18:2) | – | – | – | 0.643 | – | – | – |
| PC(30:0e) | – | – | – | 0.83 | – | – | – |
| PC(33:0) | – | – | – | 0.763 | – | – | – |
| PC(34:0e) | – | – | – | 0.884 | – | – | – |
| PC(34:2e) | – | – | – | 0.861 | – | – | – |
| PC(35:0) | – | – | – | 0.767 | – | – | – |
| PC(36:0e) | – | – | – | 0.864 | – | – | – |
| PC(36:4e) | – | – | – | 0.912 | – | – | – |
| PC(38:6e) | – | – | – | 0.839 | – | – | – |
| PC(40:3p) | – | – | – | 0.953 | – | – | – |
| PC(40:5e) | – | – | – | 0.894 | – | – | – |
| PC(40:6e) | – | – | – | 0.816 | – | – | – |
| PC(40:7p) | – | – | – | 0.804 | – | – | – |
| PC(42:1) | – | – | – | 0.781 | – | – | – |
| PC(42:3p) | – | – | – | 0.902 | – | – | – |
| PC(44:6e) | – | – | – | 0.877 | – | – | – |
| PC(44:7e) | – | – | – | 0.821 | – | – | – |
| PE(16:0p/20:4) | – | – | – | 0.868 | – | – | – |
| PE(16:0p/22:4) | – | – | – | 0.909 | – | – | – |
| PE(18:0p/18:1) | – | – | – | 0.721 | – | – | – |
| PE(18:1p/18:1) | – | – | – | 0.714 | – | – | – |
| phSM(d34:2) | – | – | – | 0.813 | – | – | – |
| phSM(d42:1) | – | – | – | 0.673 | – | – | – |
| PI(18:0/20:3) | – | – | – | 0.763 | – | – | – |
| SM(d32:0) | – | – | – | 0.817 | – | – | – |
| TG(18:2/18:2/22:6) | – | – | – | 0.784 | – | – | – |
| TG(18:3/18:2/20:4) | – | – | – | 0.716 | – | – | – |
Paper graphical summary