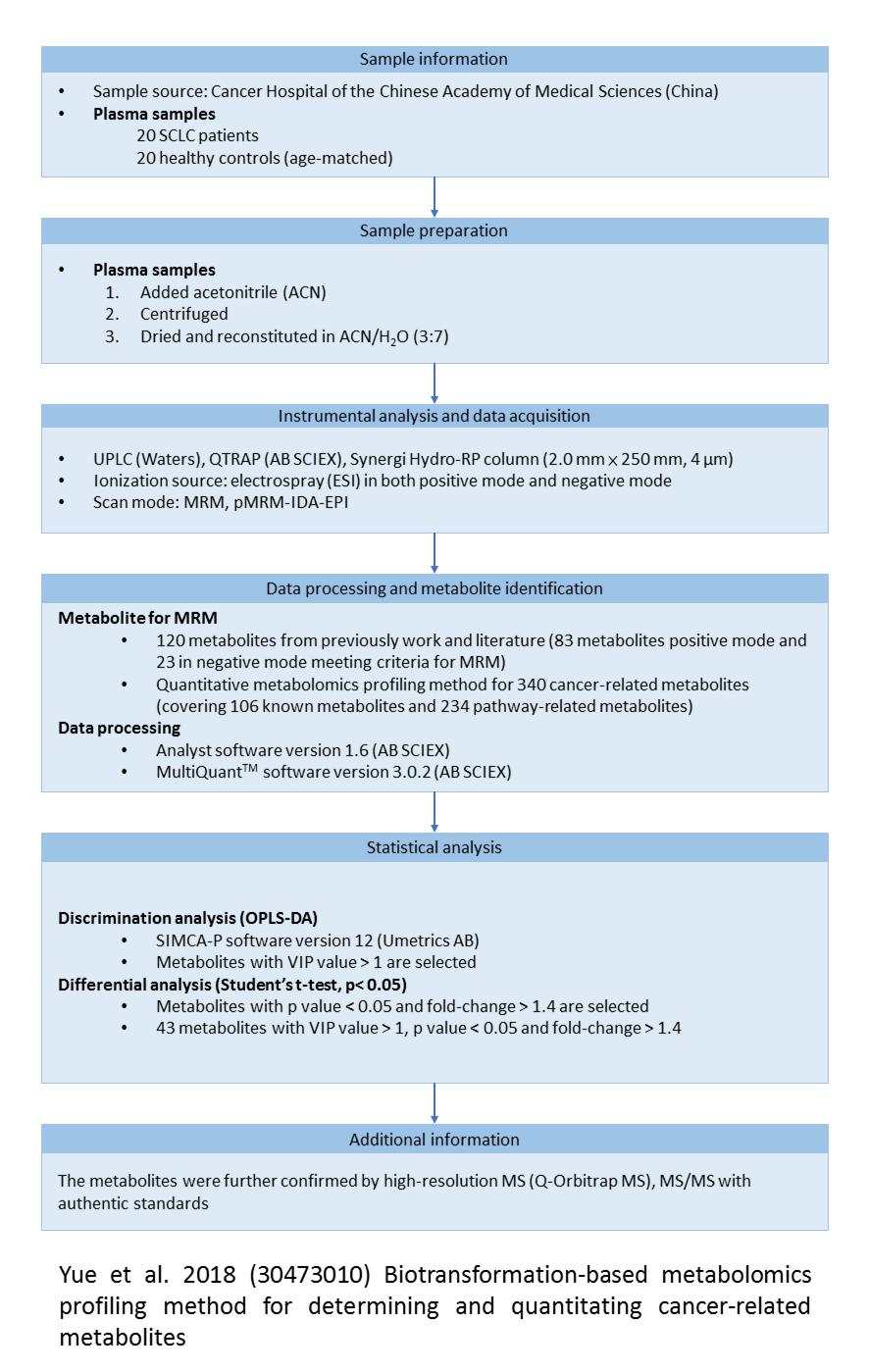
Showing paper detailed information
Citation Information
Mol Biosyst. 2013 Sep;9(9):2370-8. doi: 10.1039/c3mb70138g.
Exploratory investigation of plasma metabolomics in human lung adenocarcinoma.
Wen T, Gao L, Wen Z, Wu C, Tan CS, Toh WZ, Ong CN.
Analytical methods | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Chromatography | GC | LC | |
| Ion source | EI | ESI | |
| Positive/Negative mode | – | – | |
| Mass analyzer | – | Q-TOF | |
| Identification level | – | MS/MS | |
Sample information | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Country | China | China | |
| Specimen | plasma | plasma | |
| Marker function | diagnosis | diagnosis | |
| Participants(Case) | Cancer type | adenocarcinoma | adenocarcinoma |
| Stage | I | I | |
| Number | 31 | 31 | |
| Gender (M,F) | 15, 16 | 15, 16 | |
| Mean age (range) (M,F) | median: 63 (40-81) | median: 63 (40-81) | |
| Smoking status | smoker, non-smoker | smoker, non-smoker | |
| Participants(Control) | Type | healthy | healthy |
| Number | 28.0 | 28.0 | |
| Gender (M,F) | 20, 8 | 20, 8 | |
| Mean age (range) (M,F) | median: 37 (29-50) | median: 37 (29-50) | |
| Smoking status | smoker, non-smoker | smoker, non-smoker | |
Data processing and metabolite identification | |
|---|---|
| Data processing software | MassHunter, Mass Profiler Professional software (Agilent) |
| Database search | NIST 08, HMDB, METLIN, LIPID MAPS |
Statistics and concentration information | |
|---|---|
| Differential analysis method | Mann–Whitney–Wilcoxon test, OPLS-DA |
| Classification method | ROC curve analysis |
| Survival analysis method | – |
Metabolites identified in the paper
| Metabolite | Comparative study | Author-emphasized biomarkers | Mean concentration (case) | Mean concentration (control) | Fold change (case/control) | P-value | FDR | VIP |
|---|---|---|---|---|---|---|---|---|
| 5-hydroxytryptophan | – | – | – | 0.371130892657262 | 0.00772 | – | 1.19 | |
| glutamine | – | – | – | – | 0.438302860658018 | 0.00652 | – | 1.4 |
| threonine | – | – | – | – | 0.303548721098762 | 0.0045 | – | 1.44 |
| 3-hydroxybutyric acid | – | – | – | – | 0.360982298880624 | 0.00146 | – | 1.57 |
| alanine | – | – | – | – | 0.38156480224014 | 0.000712 | – | 1.39 |
| deoxycholic acid glycine conjugate | – | – | – | – | 0.351111218934499 | 0.000476 | – | 1.05 |
| N(6)-Methyllysine | – | – | – | – | 0.150725978461345 | 0.000423 | – | 1.17 |
| PE(38:6) | – | – | – | – | 0.346277367027731 | 0.000148 | – | 1.18 |
| glycine | – | – | – | – | 0.325335463860483 | 0.0000319 | – | 1.51 |
| PS(38:4) | – | – | – | – | 0.102948877158447 | 0.0000152 | – | 1.17 |
| uric acid | – | – | – | – | 0.26609254561334 | 0.0000132 | – | 1.26 |
| pipecolic acid | – | – | – | – | 0.325335463860483 | 0.0000119 | – | 1.16 |
| pregnenolone sulfate | – | – | – | – | 0.185565446328631 | 0.0000116 | – | 1.23 |
| PE(34:2) | – | – | – | – | 0.299369676154732 | 0.000000944 | – | 1.12 |
| PE(36:2) | – | – | – | – | 0.323088207659373 | 0.000000944 | – | 1.12 |
| ceramides (42:0) | – | – | – | – | 0.083042863381032 | 0.000000893 | – | 1.08 |
| PC(34:4) | – | – | – | – | 0.134903529563053 | 0.000000527 | – | 1.22 |
| L-arginine | – | – | – | – | 0.129408115480172 | 0.000000179 | – | 1.13 |
| PE(36:4) | – | – | – | – | 0.128514228332008 | 0.000000127 | – | 1.31 |
| proline | – | – | – | – | 0.145591698308557 | 0.0000000549 | – | 1.03 |
| PE(40:4) | – | – | – | – | 0.00955187716946929 | 0.0000000326 | – | 1.07 |
| palmitic acid | – | – | – | – | 4.25748072981344 | 0.0000000179 | – | 1.16 |
| N-Palmitoleoyl ethanolamide | – | – | – | – | 19.0273138400435 | 0.00000000514 | – | 1.03 |
| maltitol | – | – | – | – | 61.8199250511901 | 0.00000000435 | – | 1.25 |
| androsterone sulfate | – | – | – | – | 0.12586944375709 | 0.00000000353 | – | 1.11 |
| ubiquinol | – | – | – | – | 5.5404378724437 | 0.00000000153 | – | 1.23 |
| ubiquinone | – | – | – | – | 5.61777950295199 | 0.00000000115 | – | 1.29 |
| PE(38:4) | – | – | – | – | 0.102237757319723 | 0.000000000952 | – | 1.23 |
| testosterone sulfate | – | – | – | – | 0.0987551639829221 | 0.000000000486 | – | 1.22 |
| linoleic acid | – | – | – | – | 7.46426393229446 | 0.000000000441 | – | 1.25 |
| bilirubin | – | – | – | – | 290.018274635724 | 0.000000000315 | – | 1.23 |
| oleic acid | – | – | – | – | 15.6707247613908 | 0.000000000245 | – | 1.28 |
| PE(36:1) | – | – | – | – | 0.0563281539131769 | 0.000000000144 | – | 1.36 |
| PE(40:5) | – | – | – | – | 0.0131390064883393 | 0.0000000000809 | – | 1.3 |
| amylose | – | – | – | – | 709.176047672794 | 0.0000000000409 | – | 1.34 |
| 3,4,5-trimethoxycinnamic acid | – | – | – | – | – | 0.00000000000316 | – | 1.62 |
| 5-methoxytryptophan | – | – | – | – | – | 0.00000000000316 | – | 1.61 |
| Metabolite | Comparative study | Author-emphasized biomarkers | Cutoff value | AUROC (95%CI) | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| 5-hydroxytryptophan | – | – | – | 0.7 | – | – | – |
| glutamine | – | – | – | 0.71 | – | – | – |
| threonine | – | – | – | 0.71 | – | – | – |
| 3-hydroxybutyric acid | – | – | – | 0.74 | – | – | – |
| alanine | – | – | – | 0.76 | – | – | – |
| deoxycholic acid glycine conjugate | – | – | – | 0.76 | – | – | – |
| N(6)-Methyllysine | – | – | – | 0.75 | – | – | – |
| PE(38:6) | – | – | – | 0.79 | – | – | – |
| glycine | – | – | – | 0.82 | – | – | – |
| PS(38:4) | – | – | – | 0.81 | – | – | – |
| uric acid | – | – | – | 0.83 | – | – | – |
| pipecolic acid | – | – | – | 0.83 | – | – | – |
| pregnenolone sulfate | – | – | – | 0.83 | – | – | – |
| PE(34:2) | – | – | – | 0.87 | – | – | – |
| PE(36:2) | – | – | – | 0.87 | – | – | – |
| ceramides (42:0) | – | – | – | 0.86 | – | – | – |
| PC(34:4) | – | – | – | 0.85 | – | – | – |
| L-arginine | – | – | – | 0.89 | – | – | – |
| PE(36:4) | – | – | – | 0.9 | – | – | – |
| proline | – | – | – | 0.9 | – | – | – |
| PE(40:4) | – | – | – | 0.9 | – | – | – |
| palmitic acid | – | – | – | 0.93 | – | – | – |
| N-Palmitoleoyl ethanolamide | – | – | – | 0.93 | – | – | – |
| maltitol | – | – | – | 0.93 | – | – | – |
| androsterone sulfate | – | – | – | 0.95 | – | – | – |
| ubiquinol | – | – | – | 0.96 | – | – | – |
| ubiquinone | – | – | – | 0.96 | – | – | – |
| PE(38:4) | – | – | – | 0.96 | – | – | – |
| testosterone sulfate | – | – | – | 0.97 | – | – | – |
| linoleic acid | – | – | – | 0.97 | – | – | – |
| bilirubin | – | – | – | 0.96 | – | – | – |
| oleic acid | – | – | – | 0.98 | – | – | – |
| PE(36:1) | – | – | – | 0.97 | – | – | – |
| PE(40:5) | – | – | – | 0.98 | – | – | – |
| amylose | – | – | – | 0.98 | – | – | – |
| 3,4,5-trimethoxycinnamic acid | – | – | – | 1 | – | – | – |
| 5-methoxytryptophan | – | – | – | 1 | – | – | – |
Paper graphical summary